CRISPR Growing to be Game Changer for Tree Fruit
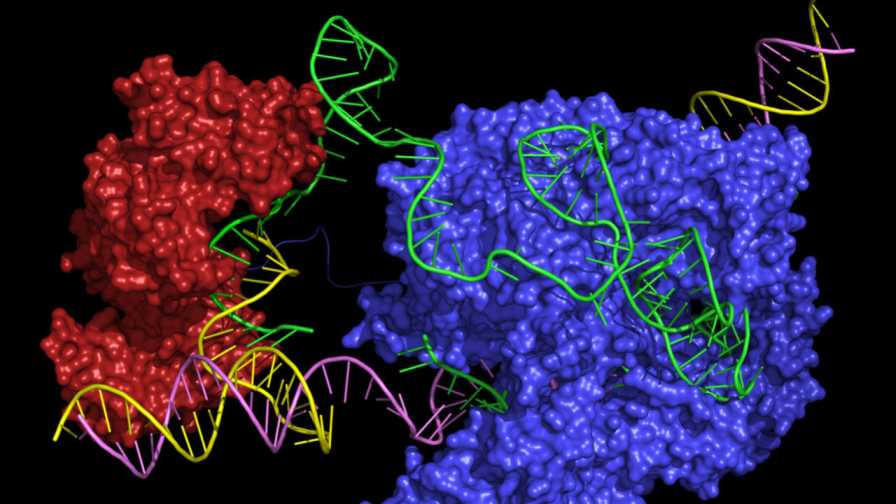
A hypothetical 3-D model of a prime editor in action. The model includes two enzymes Cas9 (blue) and reverse transcriptase (red) and a search guide, pegRNA (green), that also encodes an edit. The genomic DNA is shown in yellow and purple.
Graphic courtesy of Science magazine
There is no best, only better. This seems truly the case in the history of genome editing technology. When the CRISPR/CAS9 (clustered regularly interspaced short palindromic repeats/CRISPR associated-proteins) system was first developed in 2012-13, dubbed CRISPR 1.0, it was considered the best system ever in genome editing. But no longer.
I wrote about the CRISPR/CAS9 system previously here. In brief, it is a molecular scissor developed from a bacterial defense system that can be reprogrammed to make a cut across both DNA strains at a target genome site in animal and plant cells. Since the cut is a severe damage to DNA, it will be repaired by an innate cell DNA repair system, which is imperfect and frequently leads to knocking out genes.
Although it is still being used widely due to its effectiveness in shutting down genes of interest, CRISPR 1.0 has been exposed with critical limitations, such as an inability to precisely replace, delete, and insert a single or more predetermined DNA base in the genome, a relatively high off-target rate, and restrictions on certain genome sites that are not accessible.
To improve CRISPR 1.0, many different CRISPR/CAS9 variants have been developed and tested, including the latest one, called prime editors.
MAJOR UPGRADE
Prime editors are a major upgrade of CRISPR 1.0 that can eliminate or minimize the limitations aforementioned, representing an enormous breakthrough in CRISPR/CAS9-based genome editing technology. Many scientists consider prime editors a game changer in genome editing.
The development of prime editors was led by Dr. David Liu at Broad Institute of Harvard and MIT in Cambridge, MA. The relevant findings about prime editors were published in the prestigious scientific journal Nature in late 2019 (Anzalone, et al.).
According to the study, prime editing could be used to replace a target DNA base with any other single bases, to insert a DNA of one to 44 base pairs (bp), to delete a DNA of one to 80 bp, or to do any combinations of the listed three in human and mouse cells.
With such a broad scope of DNA editing capacity, prime editing was estimated to be capable of correcting up to 89% of 75,000 known DNA variants associated with human genetic diseases.
More remarkably, off-target by prime editors was not detectable in most cases studied. Under a poorest scenario, the off-target rate of prime editors was 2.2%, compared to 98.1% by CRISPR 1.0.
HOW DO THEY WORK?
Compared to CRISPR 1.0, prime editors were improved with the following three enhancements:
1. The single-guide RNA (sgRNA), the programmable component in the CRISPR/CAS9 system, was enhanced to include an RNA sequence containing desirable edits. Such enhanced sgRNA is called prime editing guide RNA (pegRNA).
2. The CAS9 enzyme was attached with another enzyme called reverse transcriptase that can reversely tran-scribe the blueprint encoded in the pegRNA into a DNA molecule.
3. The version of CAS9 enzyme used in prime editors can only nick one target strain of the DNA rather than cut both DNA strains, making prime editing a DNA surgery milder than that by CRISPR 1.0.
When the prime editing machinery was assembled and delivered into cells, the pegRNA will help take the whole system to the genome target site, similar to what the sgRNA does in CRISPR 1.0. Then, the CAS9 will nick the DNA strain targeted for editing while the other stain remains intact.
Next, the DNA nicking will enable the reverse transcriptase to directly copy the desirable edits encoded in pegRNA into a DNA sequence at the target site, which will eventually replace the original DNA bases in both strains. Therefore, prime editing is essentially a process of “search and replace” of DNA bases in the genome.
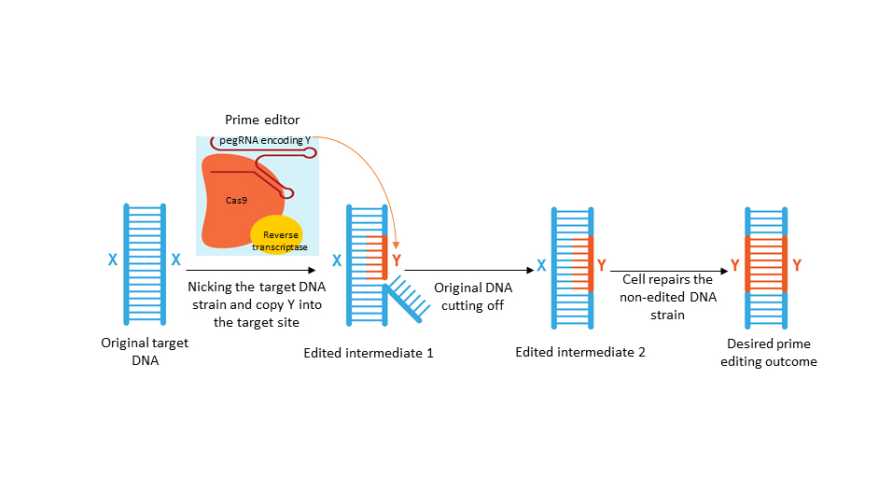
This diagram outlines the prime editing process. The prime editor complex is indicated, which includes an edit-encoding guide pegRNA and two enzymes, CAS9 (for nicking a single DNA strain) and reverse transcriptase (for reverse transcription of RNA into DNA). X=original DNA (blue); Y=edited DNA (orange).
EXPECTED OUTCOME
To date, the prime editing study has been cited by nearly 150 research articles in diverse research areas. Since such a high volume of citations was reached in just a few months, the development of prime editors clearly has sent a strong shock wave into the field of genome editing and beyond.
There is no doubt that prime editors will radically change the way how genome will be edited in the future.
However, improvement of prime editors is needed as well. One of the known issues is the large DNA size required for producing the prime editing machinery components (CAS9, reverse transcriptase, and pegRNA) in cells. This makes it difficult to deliver the DNA molecule (into cells).
First successes of using prime editors to edit genome in plant cells (rice and wheat) have been reported already, suggesting prime editing should be also extendable to genome editing in perennial woody fruit crops, such as apple and grape.
It is likely the initial applications of prime editing in woody fruit crops would be on research and on germplasm enhancement for fruit breeding. To use prime editing for direct improvement of existing fruit cultivars so that the end product is free of foreign DNA and also maintains its name recognition would still be years away due to their heterozygous genome and long juvenile phase before flowering.










